BELASCO LAB

New York University Grossman School of Medicine
Research
In all living organisms, messenger RNA (mRNA) is an indispensable intermediary that carries instructions for protein synthesis from genes to ribosomes. The universal vulnerability of mRNAs to degradation is critical for the ability of cells to adapt gene expression rapidly to their changing needs. Of the various mechanisms by which genes are regulated, mRNA decay is among the least well understood. In both eukaryotes and prokaryotes, the lifetimes of individual mRNAs can differ by as much as two orders of magnitude, with profound consequences for protein synthesis. However, despite the widespread regulatory impact of this process and its importance for human diseases ranging from microbial infection to cancer, much about it remains unexplained.
5´-terminal deprotection of mRNA
For many years it had been assumed that bacterial mRNA degradation always begins with endonucleolytic cleavage at internal sites. However, our findings have overturned that view by showing that mRNA decay is often triggered by a prior non-nucleolytic event that marks transcripts for rapid turnover: the conversion of the 5´ terminus from a triphosphate to a monophosphate. In Escherichia coli, and related organisms, this modification creates better substrates for the endonuclease RNase E, whose cleavage activity is greatly enhanced when the RNA 5´ end is monophosphorylated, whereas in Bacillus subtilis and other bacterial species that lack RNase E, it enables 5´-exonucleolytic degradation by RNase J. We have discovered and characterized a family of RNA pyrophosphohydrolases (RppH) crucial for phosphate removal from the 5´ terminus. The inability of RppH to bind monophosphorylated 5´ ends that are structurally sequestered by a stem-loop helps to explain the stabilizing influence of 5´-terminal base pairing on mRNA lifetimes in vivo. Interestingly, this master regulator of 5´-end-dependent mRNA degradation in bacteria not only catalyzes a process functionally reminiscent of eukaryotic mRNA decapping but also bears an evolutionary relationship to the eukaryotic decapping enzyme Dcp2. Current efforts are aimed at identifying the various RNA characteristics that govern the susceptibility of transcripts to RppH-dependent degradation.

Linear scanning by RNase E
The diverse lifetimes of bacterial mRNAs seem difficult to reconcile with the relaxed cleavage-site specificity of RNase E, a key regulatory endonuclease that can cut most single-stranded regions of RNA. Mounting evidence indicates that, in E. coli, rates of mRNA decay are determined not by the number or intrinsic quality of internal cleavage sites but rather by the ease with which RNase E can gain access to them. We have recently discovered that RNase E functions as a molecular zipliner, locating cleavage sites in monophosphorylated RNA by a novel mechanism that involves one-dimensional diffusion from the 5´ terminus along single-stranded RNA segments. Consequently, the rate of cleavage at those internal sites is governed not only by the ability of RNase E to initially bind the 5´ end but also by any obstacles that this endonuclease may encounter as it scans downstream. This finding has important implications for stress responses, riboswitch mechanisms, and bacterial pathogenesis. We are now investigating the mechanism of scanning and the features of RNase E that enable it to diffuse linearly on RNA.
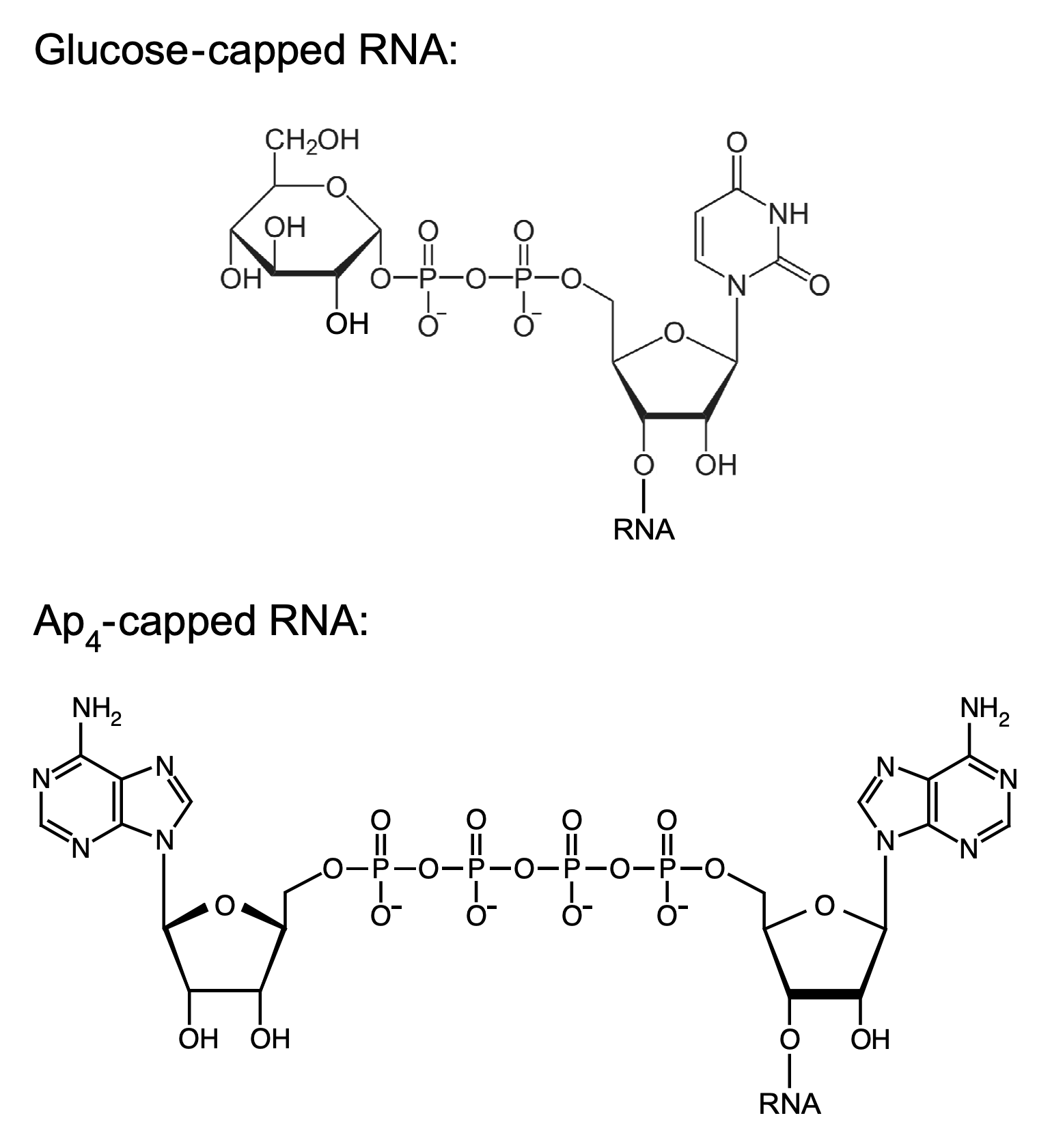
Noncanonical RNA caps
In eukaryotes, m7Gp3 caps at the 5′ end of mRNA play a crucial role in directing translation initiation and protecting mRNA from exonucleolytic degradation. By contrast, it was long assumed that bacterial RNAs are not capped. However, we and others have recently detected noncanonical caps of various kinds at the 5′ end of bacterial transcripts. In particular, the Belasco lab has discovered glucose caps and nucleoside tetraphosphate (Np4) caps on up to 75% of the 5′ ends of a variety of E. coli mRNAs and sRNAs. These caps appear to be acquired by a mechanism involving the incorporation of UDP-glucose or of a dinucleoside tetraphosphate (Np4A) into nascent transcripts by RNA polymerase during transcription initiation. Because glucose caps are resistant to removal by enzymes like RppH, they can significantly prolong the lifetime of mRNA in E. coli by protecting it from 5′-end-dependent degradation, whereas Np4 caps are readily removed by either of two pyrophosphatases, thereby triggering rapid RNA decay. Interestingly, the predominant Np4 decapping enzyme, ApaH, functions as both a sensor and an effector of disulfide stress, which inactivates ApaH and consequently impedes the degradation of Np4-capped transcripts. We are now seeking to elucidate the physiological impact of these noncanonical caps and the mechanism by which ApaH responds to stress.